SARS-CoV-2 Spike RBD Antibody Summary
Ala319-Phe541
Accession # YP_009724390.1
Applications
Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website.
Scientific Data
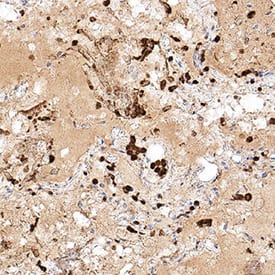 View Larger
View Larger
Detection of Spike RBD in SARS-CoV-2 infected human lung. Spike RBD was detected in immersion fixed paraffin-embedded sections of SARS-CoV-2 infected human lung using Rabbit Anti-SARS-CoV-2 Spike RBD Monoclonal Antibody (Catalog # MAB10890) at 3 µg/mL for 1 hour at room temperature followed by incubation with the Anti-Rabbit IgG VisUCyte™ HRP Polymer Antibody (Catalog # VC003). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using Antigen Retrieval Reagent-Basic (Catalog # CTS013). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to immunoreactive profiles scattered throughout the tissue. View our protocol for IHC Staining with VisUCyte HRP Polymer Detection Reagents.
Reconstitution Calculator
Preparation and Storage
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: Spike RBD
SARS-CoV-2, which causes the global pandemic coronavirus disease 2019 (Covid-19), belongs to a family of viruses known as coronaviruses that are commonly comprised of four structural proteins: Spike protein(S), Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N) (1). SARS-CoV-2 Spike Protein (S Protein) is a glycoprotein that mediates membrane fusion and viral entry. The S protein is homotrimeric, with each ~180-kDa monomer consisting of two subunits, S1 and S2 (2). In SARS-CoV-2, as with most coronaviruses, proteolytic cleavage of the S protein into two distinct peptides, S1 and S2 subunits, is required for activation. The S1 subunit is focused on attachment of the protein to the host receptor while the S2 subunit is involved with cell fusion (3-5). Based on structural biology studies, the receptor binding domain (RBD), located in the C-terminal region of S1, can be oriented either in the up/standing or down/lying state (6). The standing state is associated with higher pathogenicity and both SARS-CoV-1 and MERS can access this state due to the flexibility in their respective RBDs. A similar two-state structure and flexibility is found in the SARS-CoV-2 RBD (7). Based on amino acid (aa) sequence homology, the SARS-CoV-2 S1 subunit RBD has 73% identity with the RBD of the SARS-CoV-1 S1 RBD, but only 22% homology with the MERS S1 RBD. The low aa sequence homology is consistent with the finding that SARS and MERS bind different cellular receptors (8). The S Protein of the SARS-CoV-2 virus, like the SARS-CoV-1 counterpart, binds Angiotensin-Converting Enzyme 2 (ACE2), but with much higher affinity and faster binding kinetics (9). Before binding to the ACE2 receptor, structural analysis of the S1 trimer shows that only one of the three RBD domains in the trimeric structure is in the "up" conformation. This is an unstable and transient state that passes between trimeric subunits but is nevertheless an exposed state to be targeted for neutralizing antibody therapy (10). Polyclonal antibodies to the RBD of the SARS-CoV-2 protein have been shown to inhibit interaction with the ACE2 receptor, confirming RBD as an attractive target for vaccinations or antiviral therapy (11). There is also promising work showing that the RBD may be used to detect presence of neutralizing antibodies present in a patient's bloodstream, consistent with developed immunity after exposure to the SARS-CoV-2 virus (12). Lastly, it has been demonstrated the S Protein can invade host cells through the CD147/EMMPRIN receptor and mediate membrane fusion (13, 14).
- Wu, F. et al. (2020) Nature 579:265.
- Tortorici, M.A. and D. Veesler (2019). Adv. Virus Res. 105:93.
- Bosch, B.J. et al. (2003) J. Virol. 77:8801.
- Belouzard, S. et al. (2009) Proc. Natl. Acad. Sci. 106:5871.
- Millet, J.K. and G. R. Whittaker (2015) Virus Res. 202:120.
- Yuan, Y. et al. (2017) Nat. Commun. 8:15092.
- Walls, A.C. et al. (2010) Cell 180:281.
- Jiang, S. et al. (2020) Trends. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Ortega, J.T. et al. (2020) EXCLI J. 19:410.
- Wrapp, D. et al. (2020) Science 367:1260.
- Tai, W. et al. (2020) Cell. Mol. Immunol. https://doi.org/10.1016/j.it.2020.03.007.
- Okba, N. M. A. et al. (2020). Emerg. Infect. Dis. https://doi.org/10.3201/eid2607.200841.
- Wang, X. et al. (2020) https://doi.org/10.1038/s41423-020-0424-9.
- Wang, K. et al. (2020) bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1.
Product Datasheets
FAQs
No product specific FAQs exist for this product, however you may
View all Antibody FAQsReviews for SARS-CoV-2 Spike RBD Antibody
There are currently no reviews for this product. Be the first to review SARS-CoV-2 Spike RBD Antibody and earn rewards!
Have you used SARS-CoV-2 Spike RBD Antibody?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image

