Although HPLC and mass spectrometry can yield detailed information about glycan composition and structure, these methods often require glycan removal and labeling, which can be expensive and time-consuming.
We have developed a simple and sensitive technique for enzyme-based glycan imaging of O- and N-linked glycans. It combines the substrate specificity of glycoenzymes with specially designed azido-nucleotide-sugars and traditional click chemistry-based labeling. This technique can be used to selectively visualize the subcellular localization of glycans in cultured cells.
1. Enzymatically introduce an azido sugar onto a specific target glycan.
2. Conjugate the sugar to flourescent reporter molecule with click chemistry.
3. Image glycans by fluorescene microscopy.

Glycan Detection Reagents
See individual product pages for related enzymes and data.
Features and Benefits
Fast: No removal of glycans, mass spectrometry or HPLC
Specific: Labeling is achieved through enzymes known for selective substrate recognition
Sensitive: Labeling is through covalent bonding via click chemistry
Inexpensive: Access to mass spectrometry or HPLC is not required
Simple: Only common fluorescence equipment is needed
Visualizing Terminal Glycans
The mutant enzyme B4GalT1Y285L (or B3GALNT2) selectively installs azido GalNAc (Alexa-Fluor 555, red) on O-GlcNAc. Thus, O-GlcNAc is localized to the nucleus and cytoplasm of HUVEC cells. Nuclei are visualized using DAPI staining.
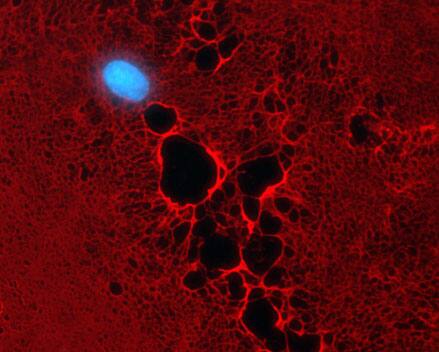
The morphology of the extracellular matrix is visualized by Heparan Sulfate (HS) staining (Alexa-Fluor 555, red). The enzymes exostosin 1 and 2 (EXT 1/2) install Azido-GlcNAc on HS. HUVEC cell nuclei are indicated by DAPI staining (blue).