 |
|
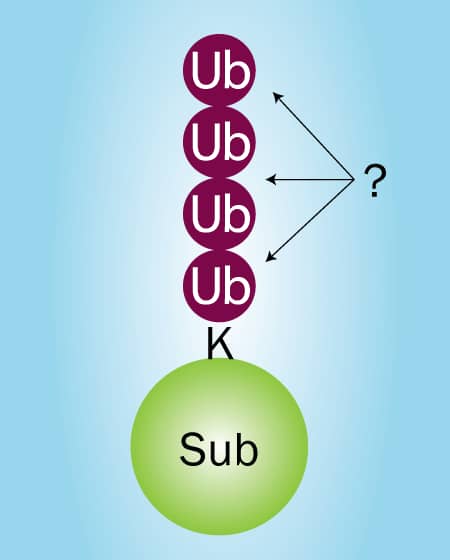
Figure 1. Eight Possible Ubiquitin Chain Linkages |
Eight residues within Ubiquitin can be utilized to form poly-Ubiquitin chains [K6, K11, K27, K29, K33, K48, K63, and Met1 (aka linear)] and linkage type directs the modified proteins to different cellular fates. Poly-Ubiquitin chains of all linkages listed above have been detected in vivo and have been shown to differentially affect many cellular processes, signaling pathways, and disease states. Explore our interactive Ubiquitination Cascade Pathway to learn more. Ubiquitin chain linkage can be determined with in vitro Ubiquitin conjugation reactions by utilizing ubiquitin lysine mutants. The protocol below describes in detail how to determine the linkage of Ubiquitin chains on your substrate of interest. Click on the links below to view listings of Boston Biochem® proteins, enzymes, and buffers.
Materials and reagents:
| Material or Reagent | Stock Concentration |
| E1 Enzyme | 5 µM |
| E2 Enzymei | 25 µM |
| E3 Ligaseii | 10 µM |
| 10X E3 Ligase Reaction Buffer | 10X - (500 mM HEPES, pH 8.0, 500 mM NaCl, 10 mM TCEP) |
| Ubiquitin | 1.17 mM (10 mg/mL) |
| Ubiquitin Mutants – Single Lysine | 1.17 mM (10 mg/mL) |
| Ubiquitin Mutants – Lysine to Arginine | 1.17 mM (10 mg/mL) |
| MgATP Solution | 100 mM |
| SDS-PAGE sample buffer – if not using reaction products for downstream applications | 2X |
| EDTA or DTT – if using reaction products for downstream applications | 500 mM (EDTA);1 M (DTT) |
| Microcentrifuge tubes | |
| Water Bath (37 °C) | |
| Western Blot Equipment |
iEach E2 enzyme functions with only a subset of E3 ligases and some E3’s are more promiscuous than others.
iiThe E3 ligase will likely need to be supplied by the user, but we do offer a small selection.
Procedure for determining ubiquitin chain linkage
 |
|
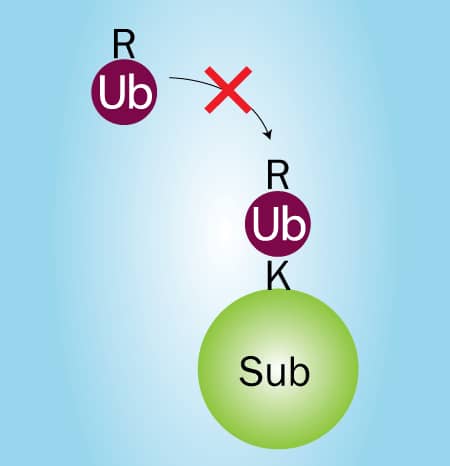
Figure 2. Lysine to Arginine Mutations Prevent Ubiquitin Chain Formation. |
Two sets of nine in vitro ubiquitin conjugation reactions will need to be performed: one set utilizing seven Ubiquitin Lysine to Arginine (K to R) Mutants followed by another set utilizing seven Ubiquitin K Only Mutants. The seven Ubiquitin K to R Mutants are used to identify the lysine or lysines being utilized for Ubiquitin chain linkage. The conjugation reaction containing the Ubiquitin K to R Mutant lacking the lysine required for chain linkage will not be able to form chains and only mono-ubiquitination will be observed by Western blot (Figure 2). For example, if Ubiquitin chains are linked via K63 then all of the conjugation reactions except the reaction containing the Ubiquitin K63R Mutant should yield Ubiquitin chains (Figure 3). If all of the conjugation reactions yield Ubiquitin chains then the chains are either linked via Met1 (linear) or they contain a mixture of linkages.*
The seven Ubiquitin K Only Mutants can then be used to verify Ubiquitin chain linkage. These Ubiquitin mutants contain only one lysine, with the remaining six mutated to arginine. Therefore, Ubiquitin chains formed with a Ubiquitin K Only Mutant must be utilizing the single lysine available for linkage. Again using Ubiquitin chains linked via K63 as an example, only the conjugation reactions containing wild type Ubiquitin and the Ubiquitin K63 Only Mutant will yield Ubiquitin chains (Figure 4).
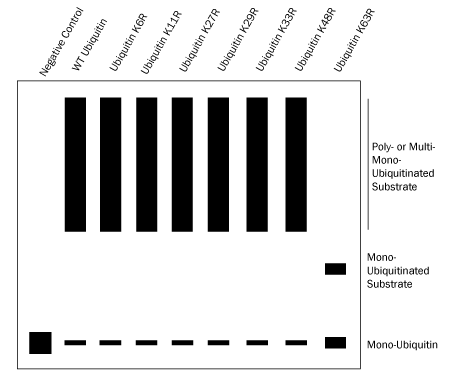 Figure 3. Determining Ubiquitin Chain Linkage Using Ubiquitin K to R Mutants. |
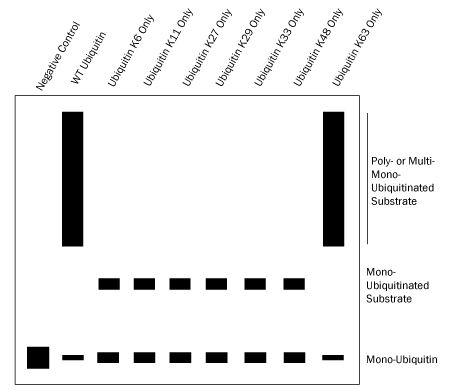 Figure 4. Verifying Ubiquitin Chain Linkage Using Ubiquitin K Only Mutants. |
*It is possible for a single Ubiquitin chain to contain multiple linkages. In this case analysis will be more complex. Contact us for technical assistance!
Procedure for 25 µL reactions (scale as needed):
- Determine Ubiquitin chain linkage. Set up nine Ubiquitin conjugation reactions: one containing wild type Ubiquitin, seven containing Ubiquitin K to R Mutants, and one negative control reaction. Combine the indicated volume of each component listed in the table below, in the order shown, in a microcentrifuge tube. Reactions 1-8 will be identical except for the Ubiquitin included:
- Reaction 1 – wild type Ubiquitin
- Reaction 2 – Ubiquitin K6R Mutant
- Reaction 3 – Ubiquitin K11R Mutant
- Reaction 4 – Ubiquitin K27R Mutant
- Reaction 5 – Ubiquitin K29R Mutant
- Reaction 6 – Ubiquitin K33R Mutant
- Reaction 7 – Ubiquitin K48R Mutant
- Reaction 8 – Ubiquitin K63R Mutant
- Negative control – replace the MgATP Solution with dH2O.
| Reagent | Volume | Working Concentration |
| dH2O | X µL (to 25 µL; dependent on volume of substrate and E3 ligase) | N/A |
| 10X E3 Ligase Reaction Buffer | 2.5 µL | 1X - (50 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM TCEP) |
| Ubiquitin or Ubiquitin K to R Mutant | 1 µL | Approximately 100 µM |
| MgATP Solution | 2.5 µL | 10 mM |
| Substrate | X µLiii | 5-10 µM |
| E1 Enzyme | 0.5 µL | 100 nM |
| E2 Enzyme | 1 µL | 1 µM |
| E3 Ligase | X µLiv | 1 µM |
iiiThe volume needed will depend on the stock concentration of your substrate.
ivThe volume needed will depend on the stock concentration of your E3 ligase.
- Incubate reactions 1-8 and the negative control reaction in a 37 °C water bath for 30-60 minutes.
- Terminate the reactions. See the table below for the appropriate method of termination.
| Are you using reaction products for downstream enzymatic applications? | Termination method | Volume (final concentration) |
| No | SDS-PAGE sample buffer | 25 µL (1X) |
| Yes | EDTA or DTTv | 0.5 µL EDTA (20 mM) or 1 µL DTT (100 mM) |
vEDTA and DTT are equally effective at terminating the reaction; determining which to use will depend on the intended downstream enzymatic application of the reaction products.
- Analyze Ubiquitin conjugation reactions by Western blot.
- Separate the reaction products by SDS-PAGE and transfer them to a PVDF or nitrocellulose membrane.
- Perform a Western blot using an anti-Ubiquitin antibody.
- Use figure 3 above as a guide to determine which lysine or lysines are required for Ubiquitin chain linkage.*
Having trouble analyzing the data? Contact us for technical support!
- Verify Ubiquitin chain linkage. Set up nine Ubiquitin conjugation reactions: one containing wild type Ubiquitin, seven containing Ubiquitin K Only Mutants, and one negative control reaction. Combine the indicated volume of each component listed in the table below, in the order shown, in a microcentrifuge tube. Reactions 1-8 will be identical except for the Ubiquitin included:
- Reaction 1 – wild type Ubiquitin
- Reaction 2 – Ubiquitin K6 Only Mutant
- Reaction 3 – Ubiquitin K11 Only Mutant
- Reaction 4 – Ubiquitin K27 Only Mutant
- Reaction 5 – Ubiquitin K29 Only Mutant
- Reaction 6 – Ubiquitin K33 Only Mutant
- Reaction 7 – Ubiquitin K48 Only Mutant
- Reaction 8 – Ubiquitin K63 Only Mutant
- Negative control – replace the MgATP Solution with dH2O.
| Reagent | Volume | Working Concentration |
| dH2O | X µL (to 25 µL; dependent on volume of substrate and E3 ligase) | N/A |
| 10X E3 Ligase Reaction Buffer | 2.5 µL | 1X - (50 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM TCEP) |
| Ubiquitin or Ubiquitin K Only Mutants | 1 µL | Approximately 100 µM |
| MgATP Solution | 2.5 µL | 10 mM |
| Substrate | X µLiii | 5-10 µM |
| E1 Enzyme | 0.5 µL | 100 nM |
| E2 Enzyme | 1 µL | 1 µM |
| E3 Ligase | X µLiv | 1 µM |
iiiThe volume needed will depend on the stock concentration of your substrate.
ivThe volume needed will depend on the stock concentration of your E3 ligase.
- Incubate reactions 1-8 and the negative control reaction in a 37 °C water bath for 30-60 minutes.
- Terminate the reactions. See the table below for the appropriate method of termination.
| Are you using reaction products for downstream enzymatic applications? | Termination method | Volume (final concentration) |
| No | SDS-PAGE sample buffer | 25 µL (1X) |
| Yes | EDTA or DTTv | 0.5 µL EDTA (20 mM) or 1 µL DTT (100 mM) |
vEDTA and DTT are equally effective at terminating the reaction; determining which to use will depend on the intended downstream enzymatic application of the reaction products.
- Analyze Ubiquitin conjugation reactions by Western blot.
- Separate the reaction products by SDS-PAGE and transfer them to a PVDF or nitrocellulose membrane.
- Perform a Western blot using an anti-Ubiquitin antibody.
- Use figure 4 above as a guide to verify which lysine or lysines are required for Ubiquitin chain linkage.*
*This approach to determining chain linkage using Ubiquitin mutants is powerful, but complimentary approaches may be required in certain cases.