Poly(ADP-ribose) Polymer/pADPr Summary
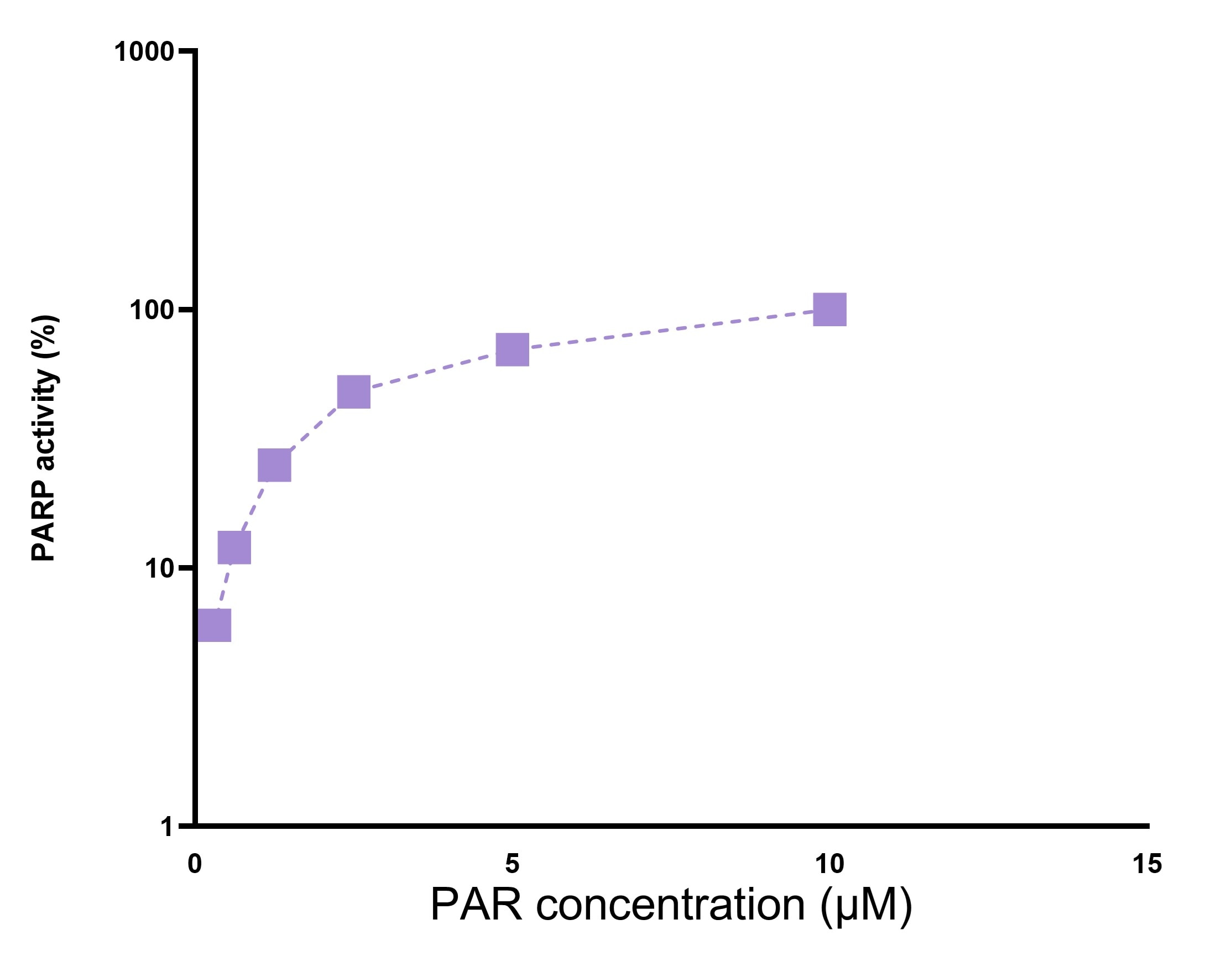
PAR Polymer was first synthesized using poly(ADP-ribose) polymerase (PARP) in the presence of NAD+, cleaved from PARP, and subsequently purified. The PAR Polymer is recognized by Trevigen's anti-PAR monoclonal and polyclonal antibodies.Specifications
Limitations
For research use only. Not for diagnostic use.
Product Datasheets
Citations for Poly(ADP-ribose) Polymer/pADPr
R&D Systems personnel manually curate a database that contains references using R&D Systems products. The data collected includes not only links to publications in PubMed, but also provides information about sample types, species, and experimental conditions.
15
Citations: Showing 1 - 10
Filter your results:
Filter by:
-
The PARP1-EXD2 axis orchestrates R-loop resolution to safeguard genome stability
Authors: Li, Z;Liu, Y;Liu, Y;Zhang, Y;Huen, MSY;Lu, H;Zhang, Z;Zhou, J;Fang, D;Liu, T;Huang, J;
Nature chemical biology 2025-06-27
-
Spermiogram, Kinetics, Flow Cytometric Characteristics and DNA Damage Degree in Boar Ejaculates: Summarization and Clustering
Authors: Ausejo-Marcos, R;Tejedor, MT;Miguel-Jiménez, S;Gómez-Giménez, B;Soriano-Úbeda, C;Mendoza, N;Vicente-Carrillo, A;Hurtado, WF;Ávila Holguín, C;Moreno, B;Falceto, MV;
Veterinary sciences 2024-09-09
-
PARG is essential for Pol?-mediated DNA end-joining by removing repressive poly-ADP-ribose marks
Authors: Vekariya, U;Minakhin, L;Chandramouly, G;Tyagi, M;Kent, T;Sullivan-Reed, K;Atkins, J;Ralph, D;Nieborowska-Skorska, M;Kukuyan, AM;Tang, HY;Pomerantz, RT;Skorski, T;
Nature communications 2024-07-11
-
SNM1A is crucial for efficient repair of complex DNA breaks in human cells
Authors: Swift, LP;Lagerholm, BC;Henderson, LR;Ratnaweera, M;Baddock, HT;Sengerova, B;Lee, S;Cruz-Migoni, A;Waithe, D;Renz, C;Ulrich, HD;Newman, JA;Schofield, CJ;McHugh, PJ;
Nature communications 2024-06-25
-
A mechanism for oxidative damage repair at gene regulatory elements
Authors: S Ray, AA Abugable, J Parker, K Liversidge, NM Palminha, C Liao, AE Acosta-Mar, CDS Souza, M Jurga, I Sudbery, SF El-Khamisy
Nature, 2022-09-28;609(7929):1038-1047. 2022-09-28
-
BTApep-TAT peptide inhibits ADP-ribosylation of BORIS to induce DNA damage in cancer
Authors: Y Zhang, M Fang, S Li, H Xu, J Ren, L Tu, B Zuo, W Yao, G Liang
Molecular Cancer, 2022-08-02;21(1):158. 2022-08-02
-
PARP1 and CHK1 coordinate PLK1 enzymatic activity during the DNA damage response to promote homologous recombination-mediated repair
Authors: B Peng, R Shi, J Bian, Y Li, P Wang, H Wang, J Liao, WG Zhu, X Xu
Nucleic Acids Research, 2021-07-21;49(13):7554-7570. 2021-07-21
-
RAD52 Adjusts Repair of Single-Strand Breaks via Reducing DNA-Damage-Promoted XRCC1/LIG3&alpha Co-localization
Authors: J Wang, YT Oh, Z Li, J Dou, S Tang, X Wang, H Wang, S Takeda, Y Wang
Cell Reports, 2021-01-12;34(2):108625. 2021-01-12
-
The Ubiquitin Ligase TRIP12 Limits PARP1 Trapping and Constrains PARP Inhibitor Efficiency
Authors: M Gatti, R Imhof, Q Huang, M Baudis, M Altmeyer
Cell Rep, 2020-08-04;32(5):107985. 2020-08-04
-
Genetically Encoded Fluorescent Sensor for Poly-ADP-Ribose
Authors: EO Serebrovsk, NM Podvalnaya, VV Dudenkova, AS Efremova, NG Gurskaya, DA Gorbachev, AV Luzhin, OL Kantidze, EV Zagaynova, SI Shram, KA Lukyanov
Int J Mol Sci, 2020-07-15;21(14):. 2020-07-15
-
Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation
Authors: AKL Leung
Trends Cell Biol., 2020-02-20;30(5):370-383. 2020-02-20
-
PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA
Authors: AS Singatulin, L Hamon, MV Sukhanova, B Desforges, V Joshi, A Bouhss, OI Lavrik, D Pastré
Cell Rep, 2019-05-07;27(6):1809-1821.e5. 2019-05-07
-
How the location of superoxide generation influences the beta-cell response to nitric oxide.
Authors: Broniowska K, Oleson B, McGraw J, Naatz A, Mathews C, Corbett J
J Biol Chem, 2015-02-03;290(12):7952-60. 2015-02-03
-
DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal.
Authors: Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P
Nucleic Acids Res, 2014-07-16;42(14):9047-62. 2014-07-16
-
PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage.
Authors: Rulten S, Rotheray A, Green R, Grundy G, Moore D, Gomez-Herreros F, Hafezparast M, Caldecott K
Nucleic Acids Res, 2013-09-18;42(1):307-14. 2013-09-18
FAQs
-
What is the concentration of this polymer in units of mass/volume?
The concentration of poly(ADP-ribose) Polymer/pADPr is determined by measuring light absorbance at 258 nm using the molar extinction coefficient of PAR. This measurement yields concentration in units of μM. The chain length of this polymer ranges between 2 and 300 ADP-ribose units. Because we are unable to evaluate the variation in polymer length and polymer branching, we are unable to calculate a concentration based on mass. As such, this product is bottled and sold by the concentration in molarity.
Reviews for Poly(ADP-ribose) Polymer/pADPr
Average Rating: 4.3 (Based on 3 Reviews)
Have you used Poly(ADP-ribose) Polymer/pADPr?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image
Filter by:
works good